As we sit at Washington Dulles airport waiting for the first leg of our trip home it seems fitting to reflect on what has been an intensely enjoyable, eye-opening, exciting, at times frustrating, but overwhelmingly awesome journey of discovery using the iPALM microscope at HHMI Janelia. Best ‘scope in the world, best three weeks of science-ing in my career.
In my post nine days ago we were reflecting on the first successful data acquired from the ‘scope. Since then, the pace ramped up rapidly.
Like with most experiments, once you get something to work you “crank the handle” and try to maximise the output. This is especially true when you have finite access to the equipment. For the final week we were joined by LaNtsandLaminin alumni Lee Troughton, who flew in from his current position at Loyola, Chicago to help with handle cranking.
We now have data from 21 regions of interest. That sounds like a tiny amount* for people used to conventional microscopy, so I thought it would help giving an idea of our days.
Our standard day started around 6am with myself or Lee starting processing the samples, antibodies on before 7am then down to Bob’s restaurant for breakfast (or outside for a “golden hour” pic of the lake+nessie (below), by 9am the coverslips for the day were ready for Anja to add imaging buffer, seal and load onto to the ‘scope.
Around 9.15am we selected the first cell to image and “raising the fork”, then Anja began the initial alignment first twiddling some tiny screw drivers to align the two objectives, correcting the final XY alignment and focusing with minute adjustments, adjusting camera/beam splitter location, then tweaking the beam splitter mirror so the correct quantity of light would reach each camera. Then waiting for the system to settle. Then repeating the whole process again. Ultimately running a calibration run and finally hitting go on the first image by maybe 11am. During this time we would be finishing up the processing from the night before.

Early lunch while the 100,000 frames (~1h) were collected in the 640nm channel, then repeating the process of alignment adjustments for the second channel. While the 150,000 images (2.5h) were collected in the 561nm laser line we ran the calibration, the initial macro and started the post-acquisition corrections for tilt, and drift.
Around 3-3.30pm we would be ready to start on the second cell. Choosing a good specimen and starting the alignment process while we also started processing the 540nm channel. By about 7pm image acquisition for both channels for cell two would be complete.
At 7-8pm Natasha would be working in the cell culture hood preparing the cells for the next day’s imaging and also maintaining the multiple other lines needed for our project. Thereafter we would get going with the processing steps and macro running of the cell two data, so it was ready to work on in the morning.
In the last few days Lee and I were trained up on how to set the imaging going. By the end of the day the system was suitably settled that the alignment steps were achievable (mostly**) by us. So, 7-9.00 …(it took us substantially longer to align than the pro) then image cell 3 channel 1 and, eventually, cell 3 channel 2. Imaging acquisition after midnight. Extra data = extra processing and by the middle of the final week Natasha would be spinning between three computers getting things going into the early hours.
We’re going home now pretty exhausted but incredibly satisfied.
I am extremely happy that I was able to bring multiple people with me. It was definitely a team effort. Natasha splitting cells every day, Tom, Lee and myself staining coverslips. Everyone pitching in to analyse the data. Without the team we’d either be zombies by now, would not have processed as much of the data and certainly wouldn’t have gathered as much data in the first place.

However, despite doing as much data processing as we possibly could during our trip, it is clear that there is so much more to do. Most of the things we’ve done so far are about quality control, but that only really gets us to the point where we can begin to answer the biology.
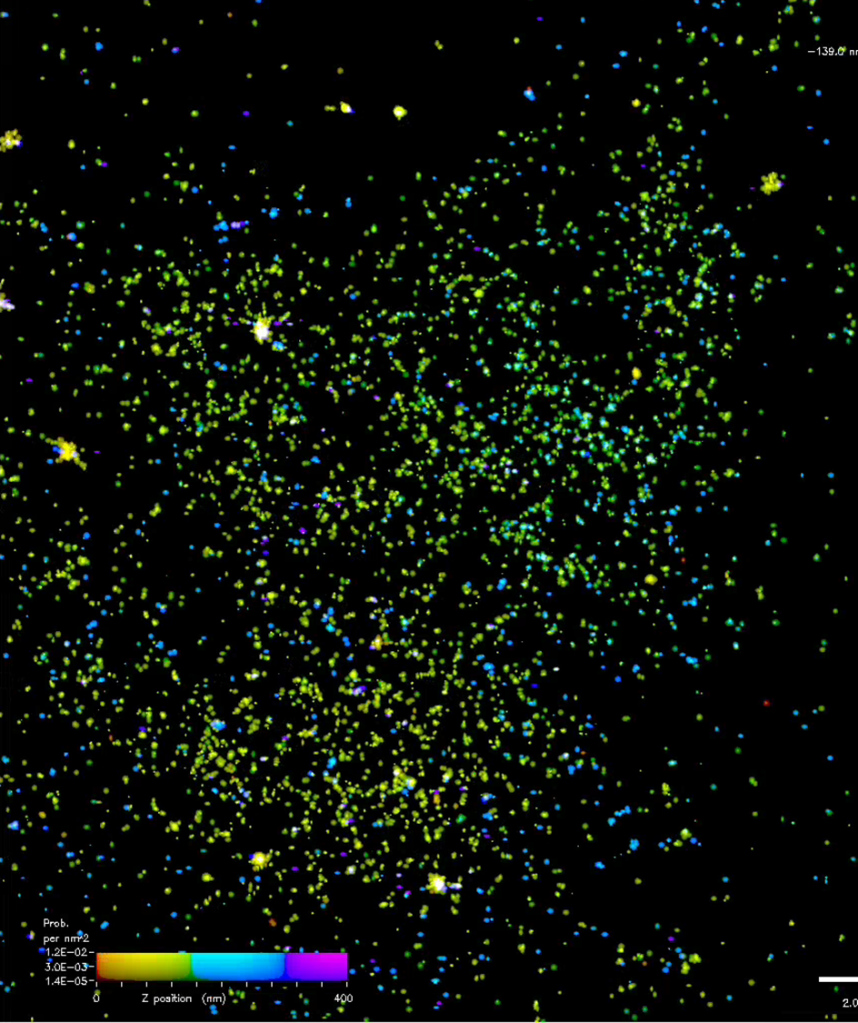
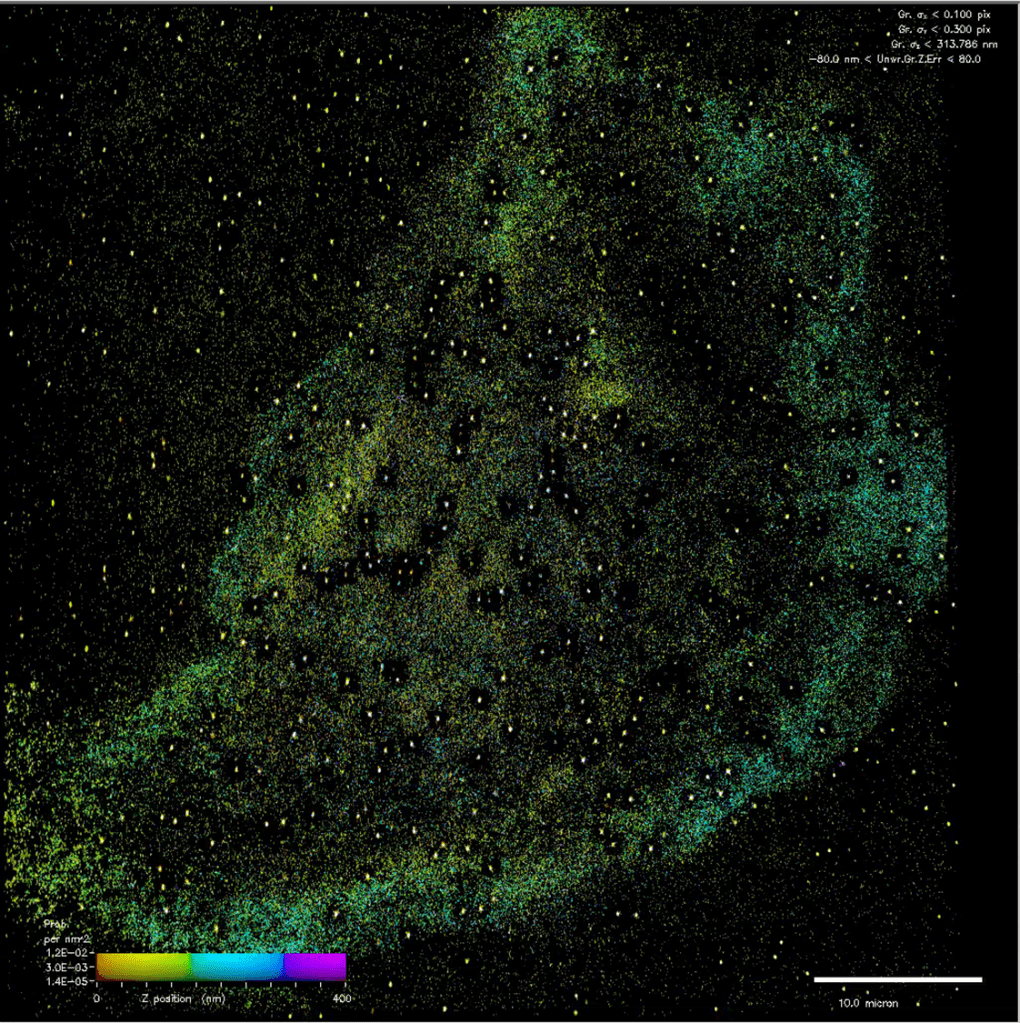
I did promise pics last time, but I think we’re going to have to wait a bit longer before they are ready. The one below and above are examples of part of the early part processed data, with different colours representing different Z heights. In the shorter term, Natasha has promised that she will write a post about her time on this trip, so look out for that soon (Natasha’s editorial note “but not too soon”).
*21 cells is >5 million individual images, ~7TB of data.
**We required Anja to come back in to help us at 8pm the first day and a technical consultation day 2 and 3. We also created a reference video filming Anja doing all the steps. It’s not easy.



2 Comments