This week Liam Shaw’s first paper came out in BMC Research Notes:
CRISPR-Cas9-mediated labelling of the C-terminus of human laminin β1 leads to secretion inhibition
https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-020-04956-z
Every paper is a reason to celebrate; however, this one is a bittersweet affair. We set out with a pretty cool plan based on solid science but sometimes you don’t get what you hoped for, and this paper describes one of those occasions.
We love laminins in all their many forms and are always looking for better ways in which we can study them. In recent years fluorescent protein technology and genome-editing have advanced rapidly, and we saw this an opportunity to generate a new line of cells where we have tagged the endogenous laminin gene with a photo-convertible protein (dendra2). This new line would allow us to do some pretty awesome things; we could study laminin using super-resolution microscopy, we study laminin dynamics and turnover and answer some bigger questions we have in terms of how basement membrane remodelling occurs, i.e. a whole bunch of stuff that’s not been possible before and which would be lots of fun.
Liam went to work. As always, we build on what has come before, and Liam based his design for the human construct based on some really elegant studies that have been done in the nematode worm C. elegans (e.g https://www.nature.com/articles/ncb2233) and also on some work that I was involved in using adenoviral constructs to express tagged human laminins (https://www.sciencedirect.com/science/article/pii/S0945053X08000978).
From this starting point, Liam designed a construct to introduce the Dendra2 sequence into the 3’ end of LAMB1 with a little linker sequence of 15 amino acids to keep the tag away from the important parts of the coiled-coil domain. Then, using CRISPR-Cas9, he genome-edited A549 cells, a widely used lung adenocarcinoma line that expresses a lot of LM511 to introduce this change.
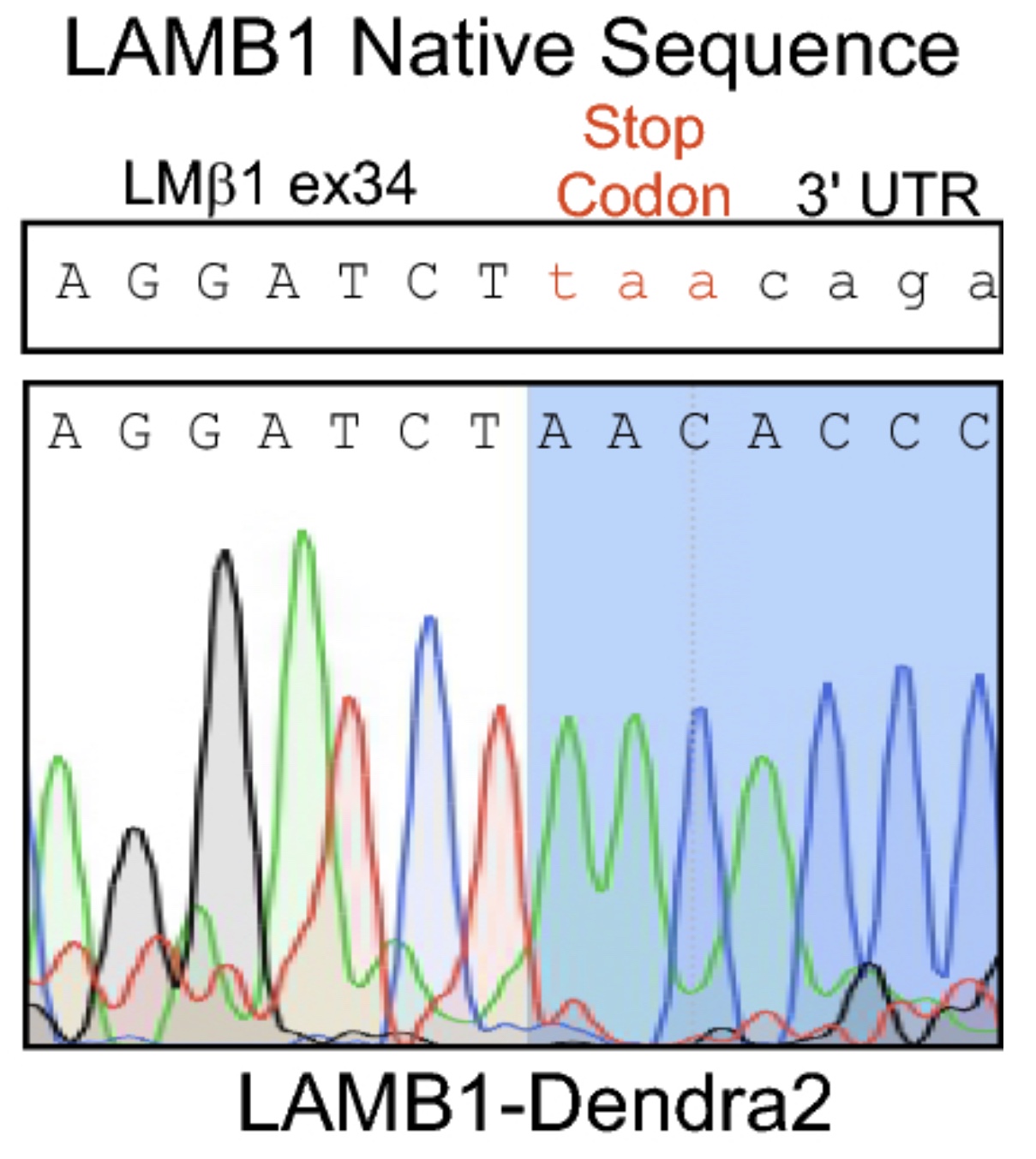
Best laid plans… gang aft agley
All was looking good. He had successfully made what he was looking for, but then the bubble burst.
Laminins are supposed to be secreted/deposited by the cells into extracellular structures that make up the road or the foundations that cells sit upon (see our intro to laminins here). However, in the genome-edited cells, It turned out that the laminin-dendra2 was getting stuck inside the cells.
Effectively this meant these cells were not useful for what we wanted them. Sad times.

Why publish something that didn’t work?
It’s not a trivial amount of work to get to this stage. Lots of cell culture, growing up hundreds of individual clones and then a load of tests to see what was going on. Literally months of work and a decent amount of money.
What hurts the most is that the plan was solid. It was that part that made us decide that this was work was valuable to the community. We do not want others to waste their effort going down a cul de sac that we have already explored. We don’t want more grant money and people’s time wasted on something that doesn’t work. We are hoping that by sharing this information, any future attempts to tag a laminin will take into account what we tried and make adjustments.
A little shout out to BMC research notes. Negative data is still data, it is still valuable. Having a home for this sort of work is really important to the scientific community.
Final comments – a valuable endeavour.
Despite not getting what we have wanted, this process has still taught us, and especially Liam, many useful things.
Liam effectively did all this pretty much by himself (the other authors are Prof Williams and myself, and we mostly just provided guidance). Liam took this project from the design of the construct used for the genome-editing, through the cell culture and electroporation, the PCR and DNA sequencing to screen clones, the blots including conditioned media precipitations, the confocal fluorescence microscopy, the flow cytometry (there’s more stuff in the paper than what I have reproduced here), and on to preparing the drafts for publishing, communicating with the journal and dealing with reviewers comments. All these skills he is putting into action in the other aspects of his PhD studies.
The rewards of this training are being felt now. He has some much more positive data coming and those data have come together a lot quicker because of honing his craft during this experience.
For anyone else pursuing an experimental research PhD, don’t ever forget that it is a learning experience and the training is an important part of the output. Not everything will work, it’s about building upon what you have learned.
So, yes, although we are disappointed that we can’t move on to some super cool imaging (yet), I’m still proud of what has been achieved and what it will mean going forward.
Check out the paper here https://bmcresnotes.biomedcentral.com/track/pdf/10.1186/s13104-020-04956-z)



Although the results of all your efforts were not what you hoped for this time, success in your future work will feel even better because of this experience. Your excellent attention to detail in your paper will be appreciated and will help other researchers Liam. Well Done.
LikeLike